Prev Post
NIE Daming

Computing is a crucial part of ZHENG Jinfang's biological research.
From Huazhong University of Science and Technology to the University of Nebraska and then to Zhejiang Lab (ZJ Lab), ZHENG Jinfang has been deeply engaged in biological research in the past decade, aiming to unravel the mysteries of the origin of life and the evolution of species using an interdisciplinary approach involving "computing + biology". Most recently, a new study by ZHENG Jinfang and his team was published in Nature Genetics, revealing the biological relationship between Zygnematophyceae and ancient plants by means of "multi-omics data + computational tools". This provides compelling evidence for the conquest of land by aquatic plants.
This year, ZHENG Jinfang joined ZJ Lab's project of open computing platform for life sciences ("life science platform"), where he focused mainly on the foundation model for single-cell omics (SCO). In his view, the "computing + data + model" approach enables automation and scale-up in biological research to improve efficiency, and can even develop "tailor-made" AI solutions to scientific problems.

Amidst towering trees, green grass and fragrant flowers, we have long been accustomed to the abundance of plant "friends" around us. However, when the Earth's geochronological clock is turned back to the Cambrian period (500 million years ago), the first lifeforms started in the ocean, and no plants yet colonized the land.
So, how did aquatic plants conquer the land? How did plants evolve from aquatic to terrestrial life? How do the environment and genetic factors interact? Urged by curiosity and these questions, ZHENG Jinfang and his team have been doing research in each of those areas.

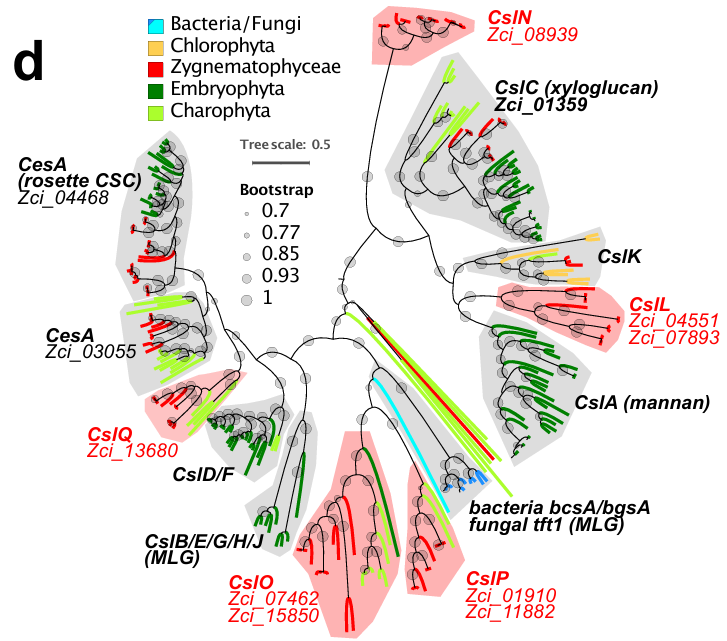
It is not easy for plants to evolve from living in the oceans to living on land, because aquatic plants had to adapt to a dry and high UV-radiation environment. In the study, ZHENG Jinfang and his team found that the amphibious Zygnematophyceae is related to land plants, and its origin may date back to remote antiquity. The team then used more than 30 open-source biocomputing tools, e.g. RAxML, to process protein sequence data of up to 16 species and 200G of transcriptomics and genomics data, and thus obtained a dendrogram showing genetically evolutionary relationships between species.
The results suggest that both Zygnematophyceae and modern plants share the cellulose synthase enzymes required for cell wall synthesis, and have the signaling network modulation mechanism necessary for adaptation to the terrestrial environment.
Since his research in computational biology, ZHENG Jinfang has been thinking about how to speed up research and offer higher-quality solutions to biological problems. Traditional biological studies involve a large number of wet lab experiments, which have higher requirements on experimental environments and subjects, and the experimental duration is also increased accordingly. Additionally, technological and methodological limitations pose challenges to large-scale data processing and complex system analysis. Even with common computational biology tools, there are still problems such as the "batch effect", background noise, and difficulty in multi-omics data integration, which can lead to different conclusions from the analysis of cells or tissues of the same kind of different organisms. Instead, foundation models could substantially improve the performance of single-cell sequencing data analysis pipelines.
Since ZJ Lab shifted its focus to intelligent computing and explored new frontiers, ZHENG Jinfang is positioned to engage in the life science platform project and explore ways and scenarios in which foundation models are used to solve life science problems after joining the Research Center for Life Sciences Computing. "We have entered an era of sci-tech innovation that is 'computing-intensive, data-driven and model-based'. As biological research evolves, computing has changed from an auxiliary tool to an important innovation engine." ZHENG Jinfang said, "Taking Zygnematophyceae as an example, it took us two to three days to process multi-omics data. If these data are processed via the life science platform, the time will be shortened to less than 5 minutes."
ZHENG Jinfang is currently involved in the SCO foundation model project. He plans to use this foundation model to process massive multi-omics data in an effort to fully demonstrate the law of cell evolution and development. "With data processing, model optimization and computational power support, the ultimate goal of AI for Science is to use foundation models to overcome specific biological problems," concluded ZHENG Jinfang. Next, he and his team will focus their research on abnormal embryonic development, blood cell function variation and immune deficiency, etc., and build and optimize platform capabilities based on data-driven models to fulfill the tremendous potential of "IT+BT" interdisciplinary methodology, thus accelerating a paradigm shift in traditional scientific research.